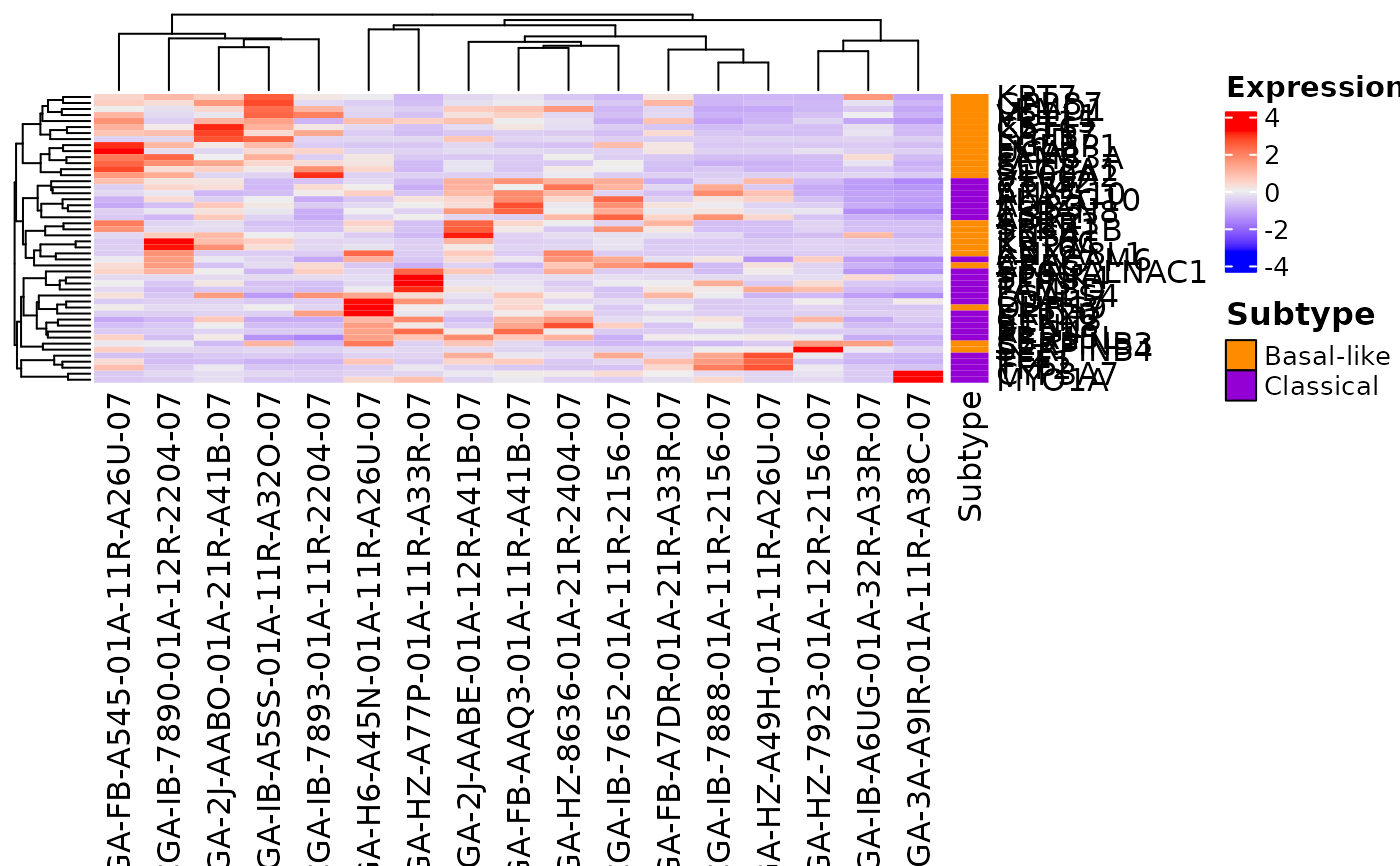
TODO
data(demo_PDAC_TCGA)Format
a data.frame containing normalized expected counts for genes
associated to Basal-like/Classical classification. The rows are containing
normalized expected counts. The data.frame
contains one column per sample in addition to a column called 'GENE' that
contains the name of the genes represented in each row.
References
The TCGA Research Network: https://www.cancer.gov/tcga
See also
createHeatmapfor creating a heating using a gene list and RNA-seq expression data.
Examples
## Load Miyabayashi et al 2020 gene list used for
## Basal-like/Classical classification
data(Miyabayashi_2020)
## Load the expression dataset
data(demo_PDAC_TCGA)
## Create a heatmap using the Miyabayashi et al 2020 gene list
createHeatmap(gene_list=Miyabayashi_2020, rna_data=demo_PDAC_TCGA,
gene_column="GENE")