Using functional enrichment results in gprofiler2 format to create an enrichment map with multiple groups from same or different enrichment analyses in an igraph format
Source:R/methodsEmap.R
createEnrichMapMultiComplexAsIgraph.RdUser selected enrichment terms are used to create an enrichment map. The selection of the term can by specifying by the source of the terms (GO:MF, REAC, TF, etc.) or by listing the selected term IDs. The map is only generated when there is at least on significant term to graph.
Usage
createEnrichMapMultiComplexAsIgraph(
gostObjectList,
queryInfo,
showCategory = 30L,
similarityCutOff = 0.2
)Arguments
- gostObjectList
a
listofgprofiler2objects that contain the results from an enrichment analysis. The list must contain at least 2 entries. The number of entries must correspond to the number of entries for thequeryListparameter.- queryInfo
a
data.framecontains one row per group being displayed. The number of rows must correspond to the number of entries for thegostObjectListparameter. The mandatory columns are:queryName: acharacterstring representing the name of the query retained for this group). The query names must exist in the associatedgostObjectListobjects and follow the same order.source: acharacterstring representing the selected source that will be used to generate the network. To hand-pick the terms to be used, "TERM_ID" should be used and the list of selected term IDs should be passed through thetermIDsparameter. The possible sources are "GO:BP" for Gene Ontology Biological Process, "GO:CC" for Gene Ontology Cellular Component, "GO:MF" for Gene Ontology Molecular Function, "KEGG" for Kegg, "REAC" for Reactome, "TF" for TRANSFAC, "MIRNA" for miRTarBase, "CORUM" for CORUM database, "HP" for Human phenotype ontology and "WP" for WikiPathways. Default: "TERM_ID".removeRoot: alogicalthat specified if the root terms of the selected source should be removed (when present).termIDs: acharacterstrings that contains the term IDS retained for the creation of the network separated by a comma ',' when the "TERM_ID" source is selected. Otherwise, it should be a empty string ("").groupName: acharacterstrings that contains the name of the group to be shown in the legend. Each group has to have a unique name.
- showCategory
a positive
integerorNULL. If ainteger, the firstnterms will be displayed. IfNULL, all terms will be displayed. Default:30L.- similarityCutOff
a positive
numeric, larger than zero and small than 1 that represent the minimum similarity level between two nodes (terms) to be linked by an edge. Default:0.2.
Value
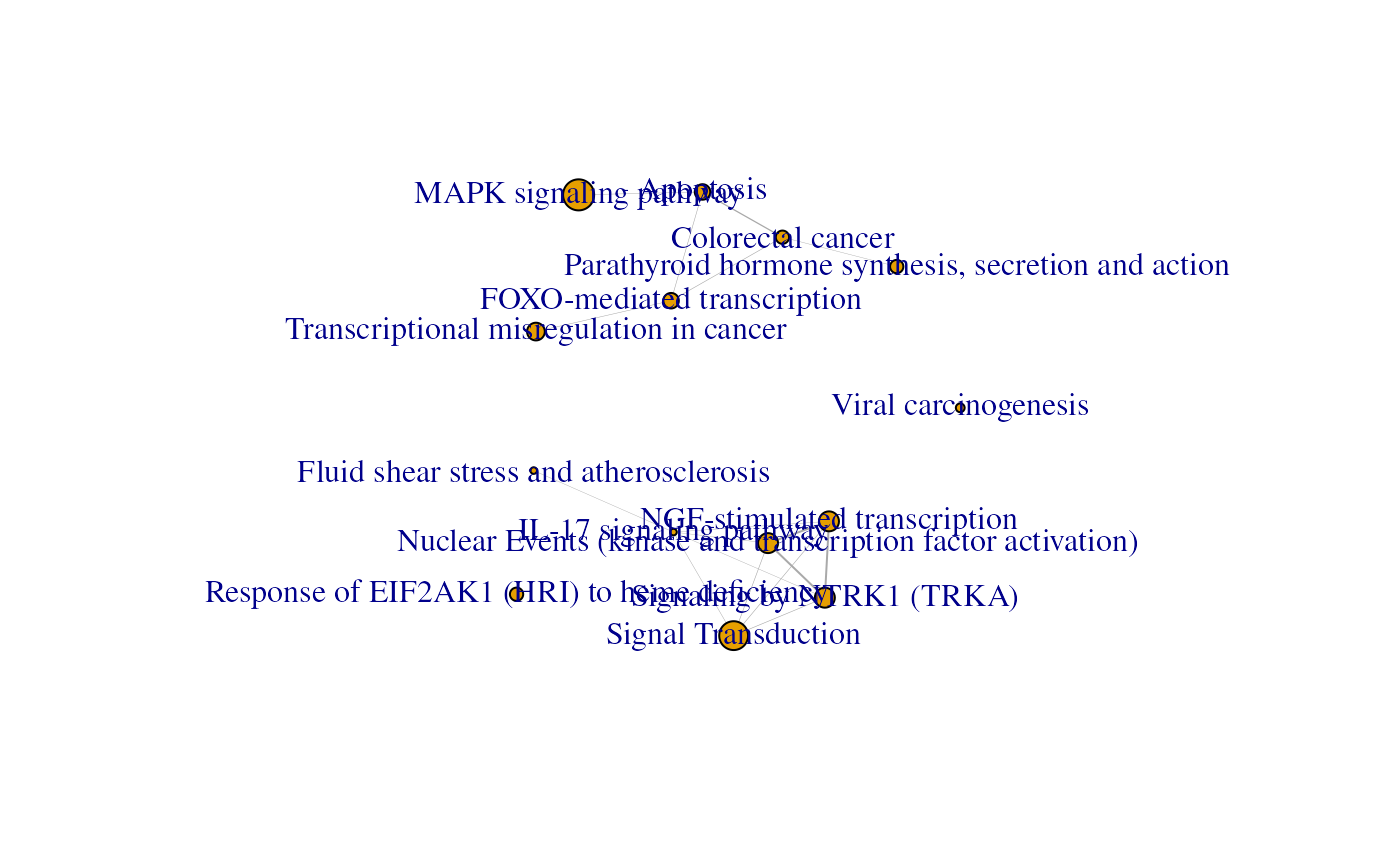
a igraph object which is the enrichment map for enrichment
results. The node have 5 attributes: "name", "size", "pie", "cluster",
and "pieName". The "name" corresponds to the term description. While the
"size" corresponds to the number of unique genes found in the specific
gene set when looking at all the experiments.
The edges have 3 attributes: "similarity", "width", and
"weight". All those 3 attributes correspond to the Jaccard coefficient.
Examples
## Loading dataset containing results from 2 enrichment analyses done with
## gprofiler2 queries
data(parentalNapaVsDMSOEnrichment)
data(rosaNapaVsDMSOEnrichment)
## The graph will be split in 4 groups
## Groups 1 and 2 are using the parental Napa vs DMSO dataset
## Group 3 and 4 are using the rosa Napa vs DMSO dataset
gostObjectList=list(parentalNapaVsDMSOEnrichment,
parentalNapaVsDMSOEnrichment, rosaNapaVsDMSOEnrichment,
rosaNapaVsDMSOEnrichment)
## Create data frame containing required information enabling the
## selection of the retained enriched terms for each enrichment analysis.
## One line per enrichment analyses present in the gostObjectList parameter
## With this data frame, the enrichment results will be split in 4 groups:
## 1) KEGG significant terms from parental napa vs DMSO (no root term)
## 2) REACTOME significant terms from parental napa vs DMSO (no root term)
## 3) KEGG significant terms from rosa napa vs DMSO (no root term)
## 4) REACTOME significant terms from rosa napa vs DMSO (no root term)
queryDataFrame <- data.frame(queryName=c("parental_napa_vs_DMSO",
"parental_napa_vs_DMSO", "rosa_napa_vs_DMSO", "rosa_napa_vs_DMSO"),
source=c("KEGG", "REAC", "KEGG", "REAC"),
removeRoot=c(TRUE, TRUE, TRUE, TRUE), termIDs=c("", "", "", ""),
groupName=c("parental - KEGG", "parental - Reactome",
"rosa - KEGG", "rosa - Reactome"), stringsAsFactors=FALSE)
## Create graph for KEGG and REACTOME significant results from
## 2 enrichment analyses in an igraph format
emap <- createEnrichMapMultiComplexAsIgraph(gostObjectList=gostObjectList,
queryInfo=queryDataFrame, showCategory=5)
if (requireNamespace("ggplot2", quietly=TRUE) &&
requireNamespace("igraph", quietly=TRUE) &&
requireNamespace("scatterpie", quietly=TRUE) &&
requireNamespace("ggtangle", quietly=TRUE) &&
requireNamespace("ggrepel", quietly=TRUE)) {
## Create a visual representation of the enrichment map
## by default
library(igraph)
plot(emap)
## Add see to reproduce the same graph
set.seed(12)
library(ggplot2)
library(ggtangle)
library(scatterpie)
library(ggrepel)
emapG <- ggplot(emap, layout=layout_with_fr) +
geom_edge(color="gray", linewidth=1)
pieInfo <- as.data.frame(do.call(rbind, V(emap)$pie))
colnames(pieInfo) <- V(emap)$pieName[[1]]
## Add information about the groups associated with each node in the
## ggplot object so that the node can be colored accordingly
for (i in seq_len(ncol(pieInfo))) {
emapG$data[colnames(pieInfo)[i]] <- pieInfo[, i]
}
## Using scatterpie, ggtangle and ggrepel to generate the graph
## geom_scatterpie() allows to have scatter pie plot
## geom_text_repel() allows to have minimum overlying terms
## coord_fixed() forces the plot to have a 1:1 aspect ratio
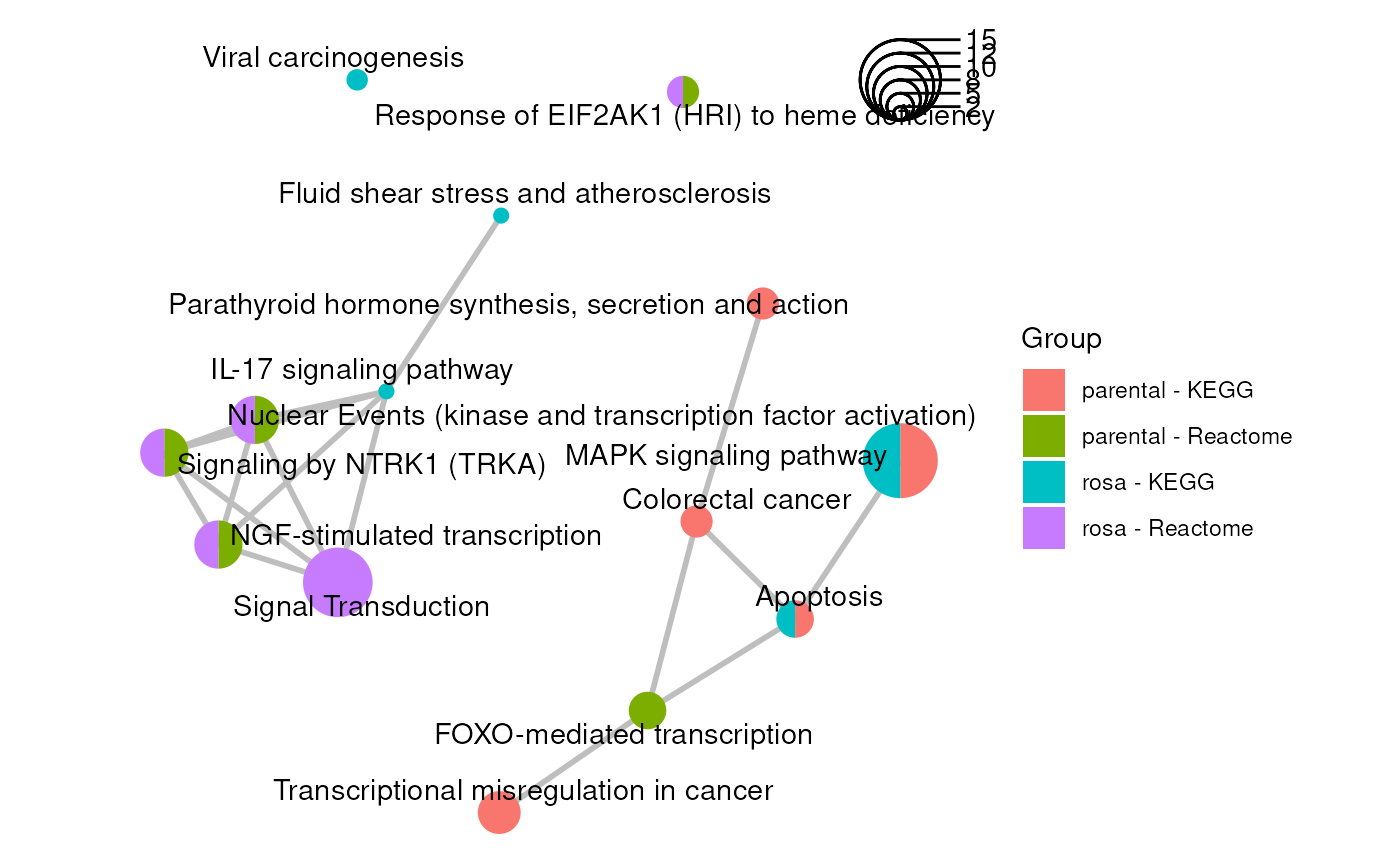
emapG +
geom_scatterpie(aes(x=x, y=y, r=size/50),
cols=c(colnames(pieInfo)), legend_name = "Group", color=NA) +
geom_scatterpie_legend(radius=emapG$data$size/50, n=4,
x=max(emapG$data$x), y=max(emapG$data$y),
labeller=function(x) {round(x*50)}, label_position="right") +
geom_text_repel(aes(x=x, y=y, label=label), max.overlaps=20) +
coord_fixed()
}